Complex dynamical biomolecular systems govern virtually all biological processes on developmental and physiological time scales. A paramount problem is to understand how structural and dynamical properties of such systems affect their roles in cellular function and dysfunction. Our group has developed network inference approaches by integrating the information from multiple types of measurement data using a variety of modeling formalisms. Such models can be used for developing optimal therapeutic strategies intended to control system behavior in disease.
B. Tercan, B. Aguilar, S. Huang, E. R. Dougherty, I. Shmulevich, “Probabilistic Boolean Networks Predict Transcription Factor Targets to Induce Transdifferentiation,” iScience, Vol. 25, No. 9, 104951, 2022.
D. L. Gibbs, B. Aguilar, V. Thorsson, A. V. Ratushny, I. Shmulevich, “Patient-specific cell communication networks associate with disease progression in cancer,” Frontiers in Genetics, Vol. 12, 667382, 2021.
D. Gibbs, I. Shmulevich, “Solving the influence maximization problem reveals regulatory organization of the yeast cell cycle,” PLoS Computational Biology, Vol. 13, No. 6, e1005591, 2017.
M. Heinäniemi, M. Nykter, R. Kramer, A. Wienecke-Baldacchino, L. Sinkkonen, J. X. Zhou, R. Kreisberg, S. A. Kauffman, S. Huang, I. Shmulevich, “Gene-pair expression signatures reveal lineage control,” Nature Methods, Vol. 10, No. 6, pp. 577-583, 2013.
A. Larjo, I. Shmulevich, H. Lähdesmäki, “Structure learning for Bayesian networks as models of biological networks,” Methods in Molecular Biology, Vol. 939, pp. 35-45, 2013.
H. Mirzaei, T. A. Knijnenburg, B. Kim, M. Robinson, P. Picotti, G. W. Carter, S. Li, D. J. Dilworth, J. K. Eng, J. D. Aitchison, I. Shmulevich, T. Galitski, R. Aebersold, J. Ranish, “Systematic measurement of transcription factor-DNA interactions by SRM mass spectrometry identifies candidate gene regulatory proteins,” Proceedings of the National Academy of Sciences of the USA, Vol. 110, No. 9, pp. 3645-3650, 2013.
S. A. Ramsey, T. A. Knijnenburg, K. A. Kennedy, D. E. Zak, M. Gilchrist, E. S. Gold, C. D. Johnson, A. Lampano, V. Litvak, G. Navarro, T. Stolyar, A. Aderem, I. Shmulevich, “Genome-wide histone acetylation data improve prediction of mammalian transcription factor binding sites,” Bioinformatics, Vol. 26, No. 17, pp. 2071-2075, 2010.
I. Shmulevich and E. R. Dougherty, Probabilistic Boolean Networks: The Modeling and Control of Gene Regulatory Networks, SIAM Press, 2009.
V. Litvak, S. Ramsey, A. Rust, D. Zak, K. Kennedy, A. Lampano, M. Nykter, I. Shmulevich, A. Aderem, “Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals,” Nature Immunology, Vol. 10, No. 4, pp. 437-43, 2009.
M. Nykter, H. Lähdesmäki, A. Rust, V. Thorsson, I. Shmulevich, “A data integration framework for prediction of transcription factor targets: a BCL6 case study,” Annals of the New York Academy of Sciences, Vol. 1158, pp. 205-214, 2009.
I. Shmulevich, J. D. Aitchison, “Deterministic and stochastic models of genetic regulatory networks,” Methods in Enzymology, Vol. 467, pp. 335-356, 2009.
A. C. Huang, L. Hu, S.A. Kauffman, W. Zhang, I. Shmulevich, “Using Cell Fate Attractors to Uncover Transcriptional Regulation of HL60 Neutrophil Differentiation,” BMC Systems Biology, Vol. 3, No. 20, 2009.
W. Liu, H. Lähdesmäki, E. R. Dougherty, I. Shmulevich, “Inference of Boolean Networks using Sensitivity Regularization,” EURASIP Journal on Bioinformatics and Systems Biology, Vol. 2008, Article ID 780541, 12 pages, 2008.
S. A. Ramsey, S. L. Klemm, D. E. Zak, K. A. Kennedy, V. Thorsson, B. Li, M. Gilchrist, E. Gold, C. D. Johnson, V. Litvak, G. Navarro, J. C. Roach, C. M. Rosenberger, A. G. Rust, N. Yudkovsky, A. Aderem, I. Shmulevich, “Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics,” PLoS Computational Biology, Vol. 4, No. 3, e1000021, 2008.
H. Lähdesmäki, A. G. Rust, I. Shmulevich, “Probabilistic inference of transcription factor binding from multiple data sources,” PLoS ONE, Vol. 3, No. 3, e1820, 2008.
H. Lähdesmäki, I. Shmulevich, “Learning the Structure of Dynamic Bayesian Networks from Time Series and Steady State Measurements,” Machine Learning, Vol. 71, pp.185-217, 2008.
N. D. Price, I. Shmulevich, “Biochemical and statistical network models for systems biology,” Current Opinion in Biotechnology, Vol. 18, pp. 365-370, 2007.
H. Lähdesmäki, S. Hautaniemi, I. Shmulevich, O. Yli-Harja, “Relationships Between Probabilistic Boolean Networks and Dynamic Bayesian Networks as Models of Gene Regulatory Networks,” Signal Processing, Vol. 86, No. 4, pp. 814-834, 2006.
M. Brun, E. R. Dougherty, I. Shmulevich, “Steady-State Probabilities for Attractors in Probabilistic Boolean Networks,” Signal Processing, Vol. 85, No. 4, pp. 1993-2013, 2005.
R. F. Hashimoto, S. Kim, I. Shmulevich, W. Zhang, M. L. Bittner, E. R. Dougherty, “Growing genetic regulatory networks from seed genes,” Bioinformatics, Vol. 20, No. 8, pp. 1241-1247, 2004.
I. Shmulevich, H. Lähdesmäki, E. R. Dougherty, J. Astola, W. Zhang, “The role of certain Post classes in Boolean network models of genetic networks,” Proceedings of the National Academy of Sciences of the USA, Vol. 100, No. 19, pp. 10734-10739, 2003.
I. Shmulevich, I. Gluhovsky, R. Hashimoto, E. R. Dougherty, and W. Zhang, “Steady-State Analysis of Genetic Regulatory Networks Modeled by Probabilistic Boolean Networks,” Comparative and Functional Genomics, Vol. 4, No. 6, pp. 601-608, 2003.
E. R. Dougherty and I. Shmulevich, “Mappings Between Probabilistic Boolean Networks,” Signal Processing, Vol. 83, No. 4, pp. 799-809, 2003.
H. Lähdesmäki, I. Shmulevich, and O. Yli-Harja, “On Learning Gene Regulatory Networks Under the Boolean Network Model,” Machine Learning, Vol. 52, pp. 147-167, 2003.
I. Shmulevich, E.R. Dougherty, and W. Zhang, “From Boolean to probabilistic Boolean networks as models of genetic regulatory networks,” Proceedings of the IEEE, Vol. 90, No. 11, pp. 1778-1792, 2002.
I. Shmulevich, E. R. Dougherty, W. Zhang, “Gene Perturbation and Intervention in Probabilistic Boolean Networks,” Bioinformatics, Vol. 18, No. 10, pp. 1319-1331, 2002.
I. Shmulevich, E.R. Dougherty, and W. Zhang, “Control of stationary behavior in Probabilistic Boolean Networks by means of structural intervention,” Journal of Biological Systems, Vol. 10, No. 4, pp. 431-445, 2002.
I. Shmulevich, E. R. Dougherty, S. Kim, W. Zhang, “Probabilistic Boolean Networks: A Rule-based Uncertainty Model for Gene Regulatory Networks,” Bioinformatics, Vol. 18, No. 2, pp. 261-274, 2002.
Featured Projects
-
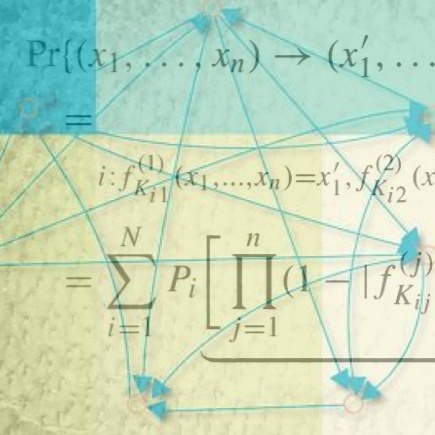
Probabilistic Boolean Networks
Probabilistic Boolean Networks are a class of models of genetic regulatory networks.
-
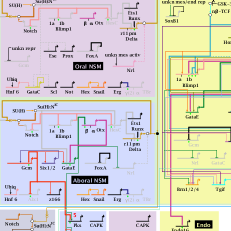
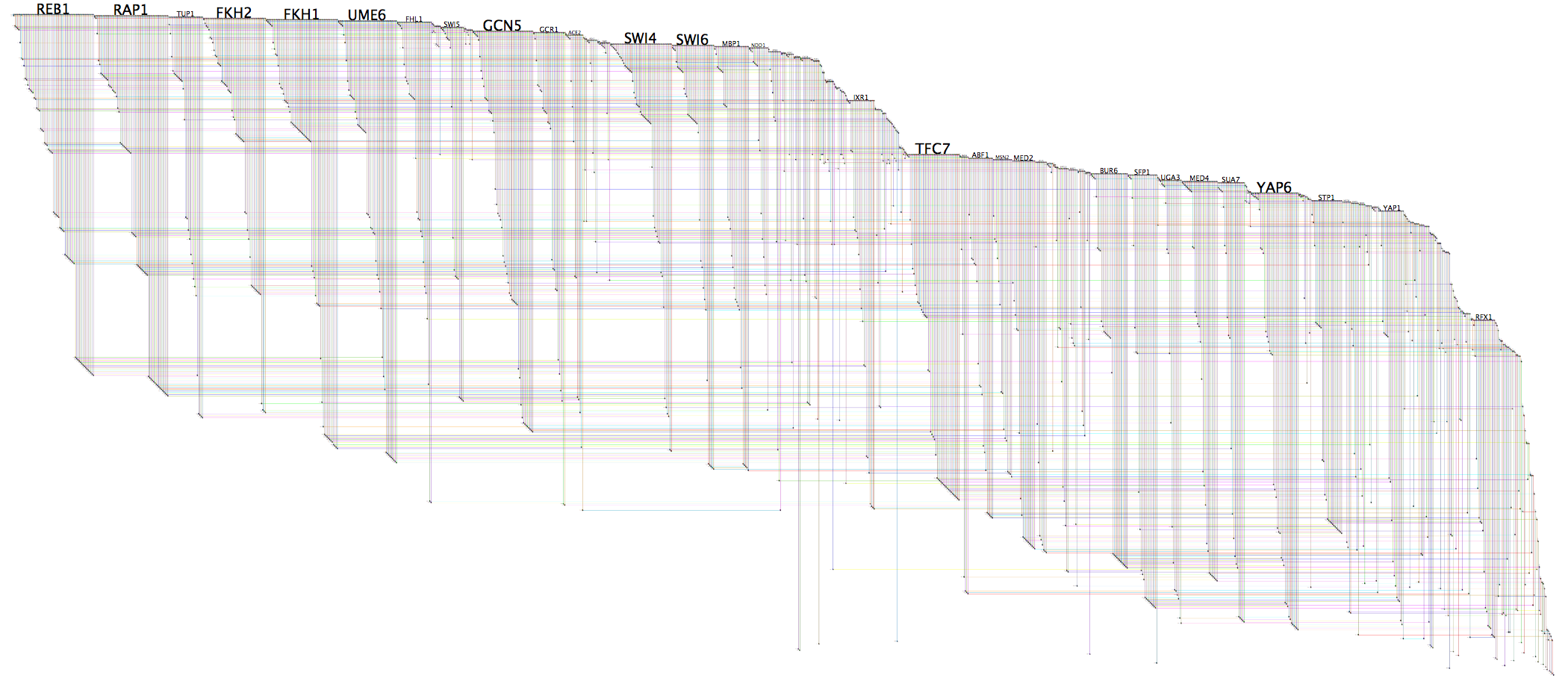
Network Visualization
Both of the network visualization tools below have been developed by our lab member Bill Longabaugh. Publications: Longabaugh, W.J.R. Combing the hairball with BioFabric: a new approach for visualization of large networks. BMC Bioinformatics, 13:275, 2012. Publications: Paquette, S.M., Leinonen, K., Longabaugh, W.J.R. BioTapestry now provides a web application and improved drawing and layout tools F1000Research 5:39, 2016. Longabaugh, W.J.R. BioTapestry: A Tool to Visualize the Dynamic Properties of Gene Regulatory…


 shmulevich.isbscience.org/research/networks/
shmulevich.isbscience.org/research/networks/