Large-scale high-throughput measurement technologies have allowed system-wide modeling and analysis of cells in health and disease. In order to be able to make reliable inferences, each type of measurement data calls for the development of appropriate statistical and computational methods.
S. A. Danziger, D. L. Gibbs, I. Shmulevich, M. McConnell, M. W. B. Trotter, F. Schmitz, D. J. Reiss, A. V. Ratushny, “ADAPTS: Automated deconvolution augmentation of profiles for tissue specific cells,” PLoS ONE, Vol. 14, No. 11, e0224693, 2019.
S. M. Reynolds, M. Miller, P. Lee, K. Leinonen, S. M. Paquette, Z. Rodebaugh, A. Hahn, D. L. Gibbs, J. Slagel, W. J. Longabaugh, V. Dhankani, M. Reyes, T. Pihl, M. Backus, M. Bookman, N. Deflaux, J. Bingham, D. Pot, I. Shmulevich, “The ISB Cancer Genomics Cloud: A Flexible Cloud-Based Platform for Cancer Genomics Research,” Cancer Research, Vol. 77, No. 21, e7-e10, 2017.
W. Poole, K. Leinonen, I. Shmulevich, T. Knijnenburg, B. Bernard, “Multiscale mutation clustering algorithm identifies pan-cancer mutational clusters associated with pathway-level changes in gene expression,” PLoS Computational Biology, Vol. 13, No. 2, e1005347, 2017.
H. T. Yang, J. H. Ju, Y. T. Wong, I. Shmulevich, J.H. Chiang, “Literature-based discovery of new candidates for drug repurposing,” Briefings in Bioinformatics, Vol. 18, No. 3, pp. 488-497, 2017.
T. A. Knijnenburg, G. W. Klau, F. Iorio, M. J. Garnett, U. McDermott, I. Shmulevich, L. F. A. Wessels, “Logic models to predict continuous outputs based on binary inputs with an application to personalized cancer therapy,” Scientific Reports, Vol. 6, 36812, 2016.
W. Poole, D. L. Gibbs, I. Shmulevich, B. Bernard, T. A. Knijnenburg, “Combining dependent P-values with an empirical adaptation of Brown’s method,” Bioinformatics, Vol. 32, No. 17, pp. i430-i436, 2016.
V. Dhankani, D. L. Gibbs, T. Knijnenburg, R. Kramer, J. Vockley, J. Niederhuber, I. Shmulevich, B. Bernard, “Using incomplete trios to boost confidence in family based association studies,” Frontiers in Genetics, Vol. 7, No. 34, 2016.
R. Bressler, R. B. Kreisberg, B. Bernard, J. E. Niederhuber, J. G. Vockley, I. Shmulevich, T. A. Knijnenburg, “CloudForest: A Scalable and Efficient Random Forest Implementation for Biological Data,” PLoS ONE, Vol. 10, No. 12, e0144820, 2015.
G. Glusman, A. Severson, V. Dhankani, M. Robinson, T. Farrah, D. Mauldin, A. B. Stittrich, S. A. Ament, J. Roach, M. Brunkow, D. Bodian, J. Vockley, I. Shmulevich, J. Niederhuber, L. Hood, “Identification of copy number variants in whole-genome data using Reference Coverage Profiles,” Frontiers in Genetics, Vol 6, No. 45, 2015.
S. Kang, S. Kahan, J. McDermott, N. Flann, I. Shmulevich, “Biocellion: Accelerating Computer Simulation of Multicellular Biological System Models,” Bioinformatics, Vol. 30, No. 21, pp. 3101-3108, 2014.
T. A. Knijnenburg, S. A. Ramsey, B. P. Berman, K. A. Kennedy, A.F.A. Smit, L. F.A. Wessels, P. W. Laird, A. Aderem, I. Shmulevich, “Multiscale representation of genomic signals,” Nature Methods, Vol. 11, pp. 689–694, 2014.
J. Lin, R. Kreisberg, A. Kallio, A. M. Dudley, M. Nykter, I. Shmulevich, P. May, R. Autio, “POMO – Plotting Omics analysis results for Multiple Organisms,” BMC Genomics, Vol. 14, No. 1, 918, 2013.
R. Bressler, J. Lin, A. Eakin, T. Robinson, R. Kreisberg, H. Rovira, T. Knijnenburg, J. Boyle, I. Shmulevich, “Fastbreak: a tool for analysis and visualization of structural variations in genomic data,” EURASIP Journal on Bioinformatics and Systems Biology, Vol. 2012(1), No. 15, 2012.
B. Bernard, V. Thorsson, H. Rovira, I. Shmulevich, “Increasing coverage of transcription factor position weight matrices through domain-level homology,” PLoS ONE, Vol. 7, No. 8, e42779, 2012.
T. Knijnenburg, O. Roda, Y. Wan, G. Nolan, J. Aitchison, I. Shmulevich, “A regression model approach to enable cell morphology correction in high-throughput flow cytometry,” Molecular Systems Biology, Vol. 7, No. 531, 2011.
T. A. Knijnenburg, J. Lin, H. Rovira, J. Boyle, I. Shmulevich, “EPEPT: A web service for enhanced P-value estimation in permutation tests,” BMC Bioinformatics, Vol. 12, No. 411, 2011.
T. Erkkilä, S. Lehmusvaara, P. Ruusuvuori, T. Visakorpi, I. Shmulevich, H. Lähdesmäki, “Probabilistic analysis of gene expression measurements from heterogeneous tissues,” Bioinformatics, Vol. 26, No. 20, pp. 2571-2577, 2010.
D. B. Burdick, C. Cavnor, J. Handcock, S. Killcoyne, J. Lin, B. Marzolf, S. A. Ramsey, H. Rovira, I. Shmulevich, J. Boyle, “SEQADAPT: An adaptable system for the tracking, storage and analysis of high throughput sequencing experiments,” BMC Bioinformatics, Vol. 11, No. 377, 2010.
T. A. Knijnenburg, L. F. A. Wessels, M. J. T. Reinders, I. Shmulevich, “Fewer permutations, more accurate P-values,” Bioinformatics, Vol. 25, No. 12, pp. i161-i168, 2009.
J. Boyle, H. Rovira, C. Cavnor, D. Burdick, S. Killcoyne, I. Shmulevich, “Adaptable Data Management for Systems Biology Investigations,” BMC Bioinformatics, Vol. 10, No. 79, 2009.
J. Boyle, C. Cavnor, S. Killcoyne, I. Shmulevich, “Systems biology driven software design for the research enterprise,” BMC Bioinformatics, Vol. 9, No. 295, 2008.
M. Korb, A. G. Rust, V. Thorsson, C. Battail, B. Li, D. Hwang, K. Kennedy, J. C. Roach, C. M. Rosenberger, M. Gilchrist, D. Zak, C. Johnson, B. Marzolf, A. Aderem, I. Shmulevich, H. Bolouri, “The Innate Immune Database (IIDB),” BMC Immunology, Vol. 9, No. 7, 2008.
R. Vêncio, I. Shmulevich, “ProbCD: enrichment analysis accounting for categorization uncertainty,” BMC Bioinformatics, Vol. 8, No. 383, 2007.
R. Vêncio, L. Varuzza, C. Pereira, H. Brentani, I. Shmulevich, “Simcluster: clustering enumeration gene expression data on the simplex space,” BMC Bioinformatics, Vol. 8, No. 246, 2007.
M. Ahdesmäki , H. Lähdesmäki, A. Gracey , I. Shmulevich and O. Yli-Harja, “Robust regression for periodicity detection in non-uniformly sampled time-course gene expression data,” BMC Bioinformatics, Vol. 8, No. 233, 2007.
I. Tabus, A. Hategan, C. Mircean, J. Rissanen, I. Shmulevich, W. Zhang, J. Astola, “Nonlinear modeling of protein expressions in protein arrays,” IEEE Transactions on Signal Processing, Vol. 54, No. 6, pp. 2394-2407, 2006.
C. Mircean, I. Shmulevich, D. Cogdell, W. Choi, Y. Jia, I. Tabus, S. R. Hamilton, W. Zhang, “Robust estimation of protein expression ratios with lysate microarray technology,” Bioinformatics, Vol. 21, No. 9, pp. 1935-1942, 2005.
H. Lähdesmäki, I. Shmulevich, V. Dunmire, O. Yli-Harja, W. Zhang, “In Silico Microdissection of Microarray Data from Heterogeneous Cell Populations,” BMC Bioinformatics 6:54, 2005.
I. Shmulevich, J. Astola, D. Cogdell, S. R. Hamilton, and W. Zhang, “Data Extraction From Composite Oligonucleotide Microarrays,” Nucleic Acids Research, Vol. 31, No. 7, e36, 2003.
I. Shmulevich, W. Zhang, “Binary Analysis and Optimization-Based Normalization of Gene Expression Data,” Bioinformatics, Vol. 18, No. 4, pp. 555-565, 2002.
Featured Projects
-
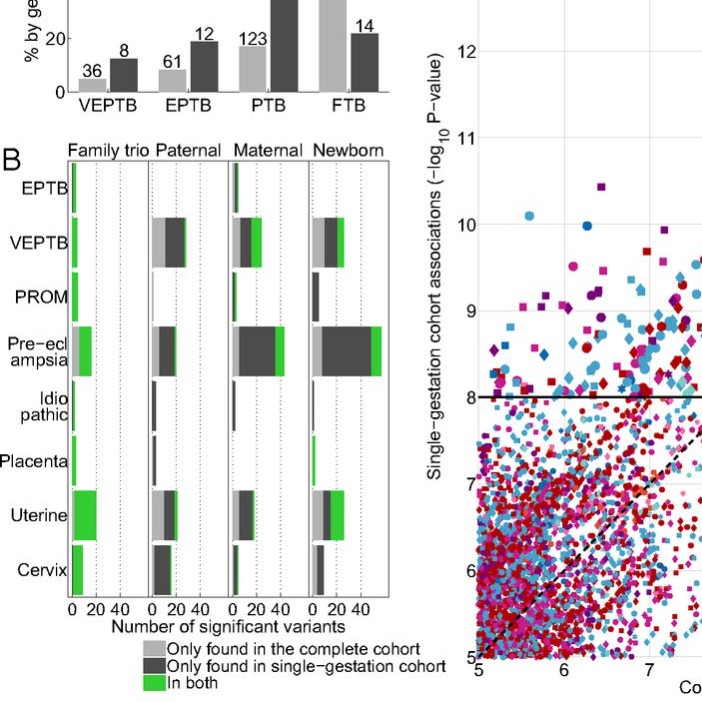
Genomic and molecular characterization of preterm birth
A family-based study of preterm birth with whole-genome sequencing, RNA sequencing, DNA methylation, and clinical data.
-

The ISB Cancer Genomics Cloud
The ISB Cancer Genomics Cloud (ISB-CGC) is democratizing access to NCI Cancer Data and coupling it with unprecedented computational power to allow researchers to explore and analyze this vast data-space.


 shmulevich.isbscience.org/research/computational-biology-tools-and-methods/
shmulevich.isbscience.org/research/computational-biology-tools-and-methods/